Multi-Class Classification (Optional)
Contents
11. Multi-Class Classification (Optional)#
11.1. Lecture Learning Objectives#
Explain components of a confusion matrix with respect to multi-class classification.
Define precision, recall, and f1-score with multi-class classification
Carry out multi-class classification using OVR and OVO strategies.
11.2. Five Minute Recap/ Lightning Questions#
What metrics is calculated using the equation \(\frac{TP}{TP + FP}\) ?
What function can be used to find the calculated values of precision, recall, and f1?
What function do we use to identify the number of false positives, false negatives and correctly identified positive and negative values?
What argument/parameter is important to use when we make our own scorer where lower values are better?
What regression metric will give funky units?
11.2.1. Some lingering questions#
What happens if we have data where there is a lot of one class and very few of another?
How do we measure precision and recall and what do our confusion matrices look like now
11.3. Multi-class classification#
Often we will come across problems where there are more than two classes to predict.
We call these multi-class problems.
Some algorithms can natively support multi-class classification, for example:
Decision Trees
\(K\)-nn
Naive Bayes
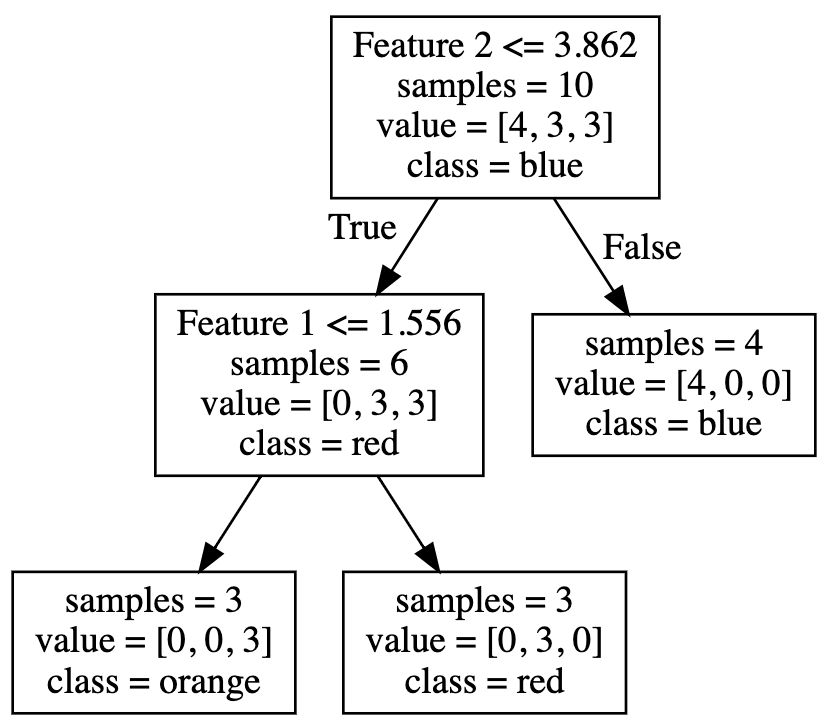
Below is an example of a Decision Tree Classifier used to classify 3 labels
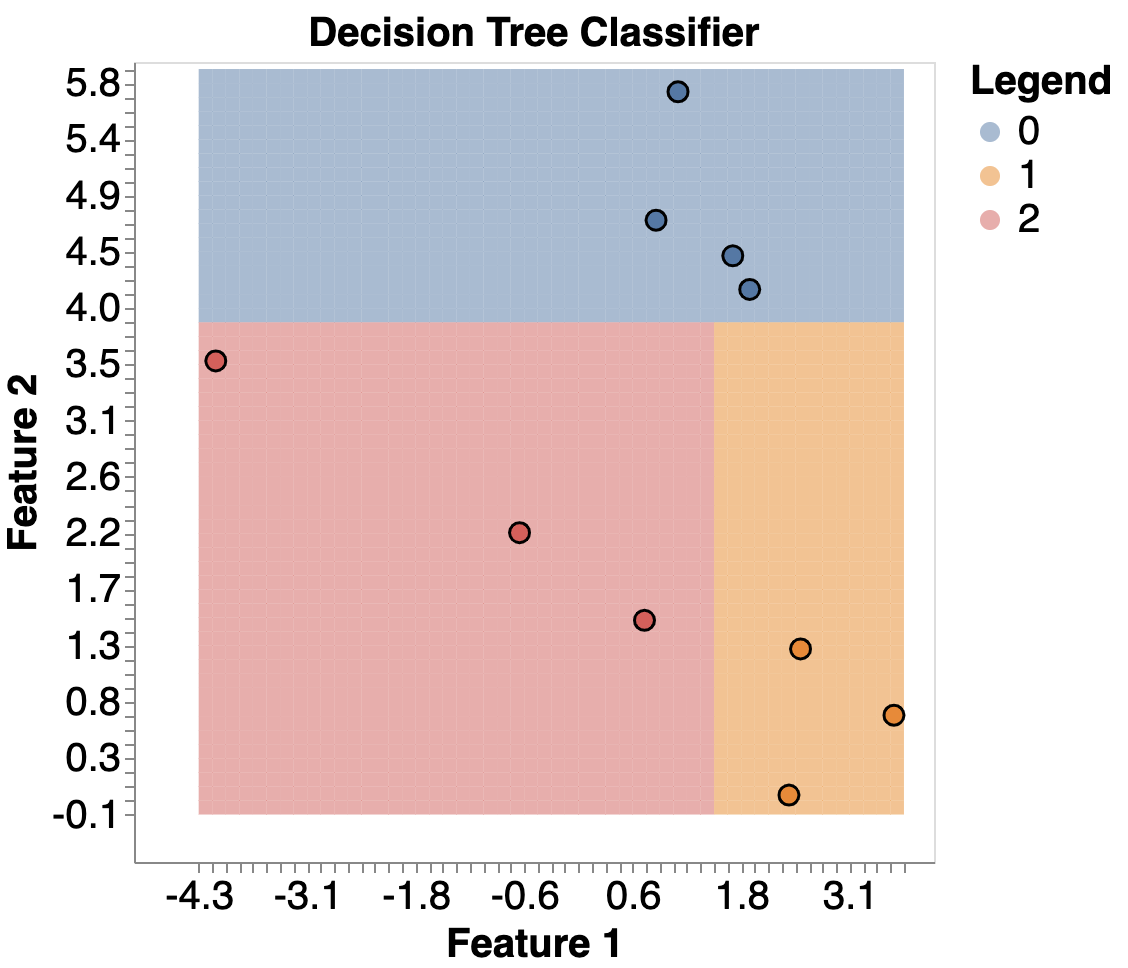
And here’s the graph:
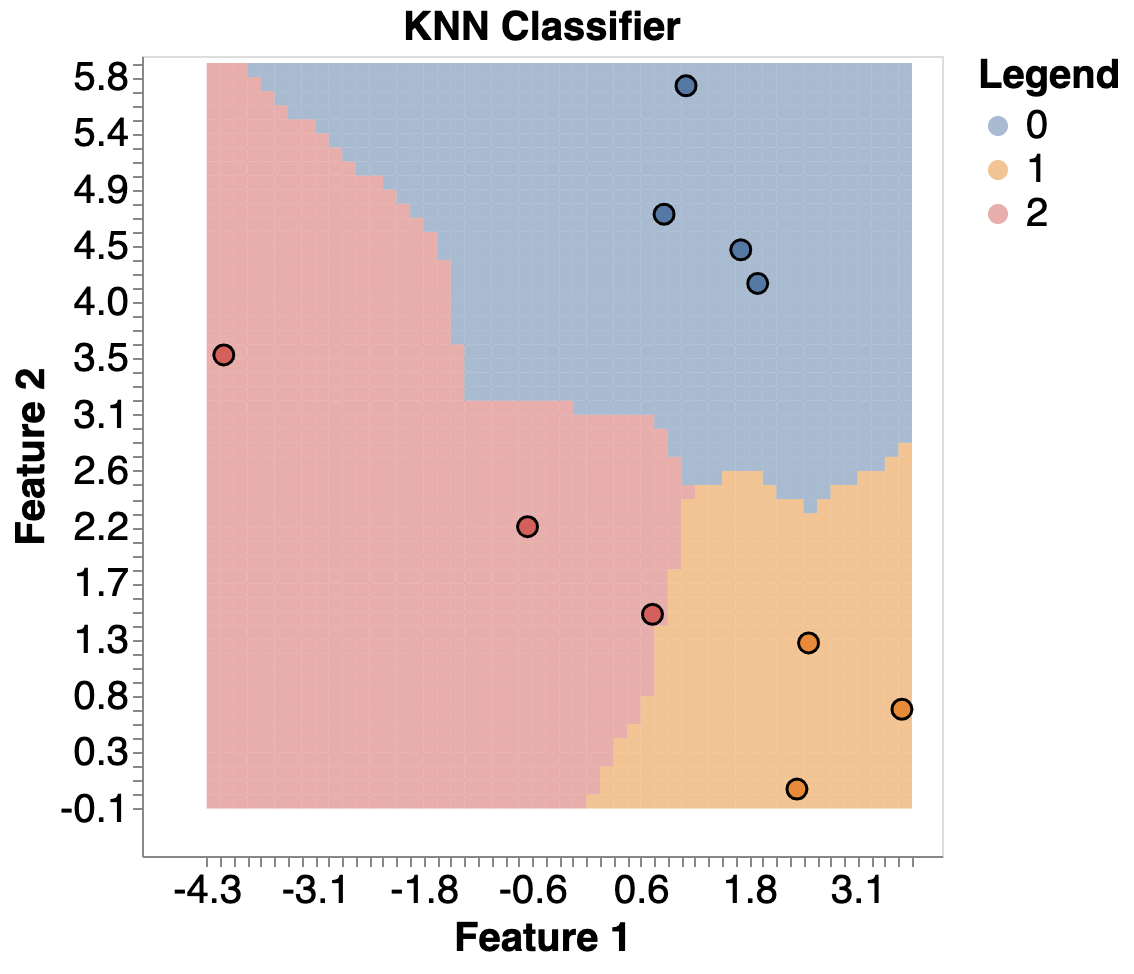
Here’s an example of KNN:
Other models, like SVMs and Logistic Regression, don’t natively support multi-class classification.
Instead, there are two common strategies to help us:
One-vs-rest
One-vs-one
11.3.1. One-vs-Rest#
(also known as one-vs-all)
It’s the default for most sklearn algorithms, e.g., LogisticRegression, SVM.
Turns \(k\)-class classification into \(k\) binary classification problems.
Builds \(k\) binary classifiers; for each classifier, the class is fitted against all the other classes.
For k classes, that means we need k models in total, e.g.:
blue vs (red & orange)
red vs (blue & orange)
orange vs (blue & red)
We use all models to make a prediction, and then choose the category with the highest prediction/probability/confidence.
You can do this yourself for any binary classifier using
OneVsRestClassifier
Here we are importing OneVsRestClassifier from sklearn.multiclass
from sklearn.multiclass import OneVsRestClassifier
We are going to use a wine dataset that has 3 different classes; 0, 1, 2 (maybe red, white and rose?)
import pandas as pd
from sklearn import datasets
from sklearn.model_selection import train_test_split
data = datasets.load_wine()
X = pd.DataFrame(data['data'], columns=data["feature_names"])
X = X[['alcohol', 'malic_acid']]
y = data['target']
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=2021)
X_train.head()
| alcohol | malic_acid | |
|---|---|---|
| 36 | 13.28 | 1.64 |
| 77 | 11.84 | 2.89 |
| 131 | 12.88 | 2.99 |
| 159 | 13.48 | 1.67 |
| 4 | 13.24 | 2.59 |
pd.DataFrame(y_train).value_counts()
1 54
0 40
2 39
dtype: int64
from sklearn.linear_model import LogisticRegression
ovr = OneVsRestClassifier(LogisticRegression(max_iter=100000))
ovr.fit(X_train, y_train)
ovr.score(X_train, y_train)
0.7819548872180451
from plot_classifier import plot_classifier
plot_classifier(X_train, y_train, ovr);
/home/joel/miniconda3/envs/bait/lib/python3.9/site-packages/sklearn/base.py:450: UserWarning: X does not have valid feature names, but LogisticRegression was fitted with feature names
warnings.warn(
/home/joel/miniconda3/envs/bait/lib/python3.9/site-packages/sklearn/base.py:450: UserWarning: X does not have valid feature names, but LogisticRegression was fitted with feature names
warnings.warn(
/home/joel/miniconda3/envs/bait/lib/python3.9/site-packages/sklearn/base.py:450: UserWarning: X does not have valid feature names, but LogisticRegression was fitted with feature names
warnings.warn(
/home/joel/miniconda3/envs/bait/lib/python3.9/site-packages/plot_classifier/__init__.py:122: UserWarning: You passed a edgecolor/edgecolors ('k') for an unfilled marker ('x'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax.scatter(X0[y==labels[2]], X1[y==labels[2]], s=60, c='k', marker='x', edgecolors='k')
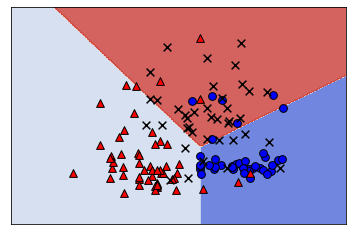
11.3.2. One-vs-One#
One-vs-One fits a model to all pairs of categories.
If there are 3 classes (“blue”, “red”, “orange”), we fit a model on:
blue vs red
blue vs orange
red vs orange
So we have 3 models in this case, or in general \(\frac{n*(n-1)}{2}\)
For 100 classes, we fit 4950 models!
All models are used during prediction and the classification with the most “votes” is predicted.
Computationally expensive, but can be good for models that scale poorly with data, because each model in OvO only uses a fraction of the dataset.
from sklearn.multiclass import OneVsOneClassifier
ovo = OneVsOneClassifier(LogisticRegression(max_iter=100000))
ovo.fit(X_train, y_train)
ovo.score(X_train, y_train)
0.8120300751879699
plot_classifier(X_train, y_train, ovo);
/home/joel/miniconda3/envs/bait/lib/python3.9/site-packages/sklearn/base.py:450: UserWarning: X does not have valid feature names, but OneVsOneClassifier was fitted with feature names
warnings.warn(
/home/joel/miniconda3/envs/bait/lib/python3.9/site-packages/plot_classifier/__init__.py:122: UserWarning: You passed a edgecolor/edgecolors ('k') for an unfilled marker ('x'). Matplotlib is ignoring the edgecolor in favor of the facecolor. This behavior may change in the future.
ax.scatter(X0[y==labels[2]], X1[y==labels[2]], s=60, c='k', marker='x', edgecolors='k')
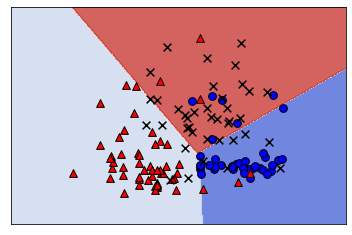
11.4. Multi-class measurements#
Similar to how we can use different classification metrics for binary classification, we can do so with multi-class too.
Let’s look at this with a larger version of this wine dataset.
data = datasets.load_wine()
X = pd.DataFrame(data['data'], columns=data["feature_names"])
y = data['target']
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=2021)
X_train.head()
| alcohol | malic_acid | ash | alcalinity_of_ash | magnesium | total_phenols | flavanoids | nonflavanoid_phenols | proanthocyanins | color_intensity | hue | od280/od315_of_diluted_wines | proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 36 | 13.28 | 1.64 | 2.84 | 15.5 | 110.0 | 2.60 | 2.68 | 0.34 | 1.36 | 4.60 | 1.09 | 2.78 | 880.0 |
| 77 | 11.84 | 2.89 | 2.23 | 18.0 | 112.0 | 1.72 | 1.32 | 0.43 | 0.95 | 2.65 | 0.96 | 2.52 | 500.0 |
| 131 | 12.88 | 2.99 | 2.40 | 20.0 | 104.0 | 1.30 | 1.22 | 0.24 | 0.83 | 5.40 | 0.74 | 1.42 | 530.0 |
| 159 | 13.48 | 1.67 | 2.64 | 22.5 | 89.0 | 2.60 | 1.10 | 0.52 | 2.29 | 11.75 | 0.57 | 1.78 | 620.0 |
| 4 | 13.24 | 2.59 | 2.87 | 21.0 | 118.0 | 2.80 | 2.69 | 0.39 | 1.82 | 4.32 | 1.04 | 2.93 | 735.0 |
X_train.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 133 entries, 36 to 116
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 alcohol 133 non-null float64
1 malic_acid 133 non-null float64
2 ash 133 non-null float64
3 alcalinity_of_ash 133 non-null float64
4 magnesium 133 non-null float64
5 total_phenols 133 non-null float64
6 flavanoids 133 non-null float64
7 nonflavanoid_phenols 133 non-null float64
8 proanthocyanins 133 non-null float64
9 color_intensity 133 non-null float64
10 hue 133 non-null float64
11 od280/od315_of_diluted_wines 133 non-null float64
12 proline 133 non-null float64
dtypes: float64(13)
memory usage: 14.5 KB
Since our data here isn’t missing any values and it’s all numeric, we can make a pipeline with just StandardScaler() and a model, we are going to use LogisticRegression.
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
pipe = make_pipeline(
(StandardScaler()),
(LogisticRegression())
)
pipe.fit(X_train,y_train);
predictions = pipe.predict(X_test)
pipe.score(X_test,y_test)
0.9333333333333333
We can predict on our test set and see that we get an accuracy of 93%.
But what does this mean for our metrics?
11.4.1. Multiclass confusion metrics#
We can still create confusion matrices but now they are greater than a 2 X 2 grid.
We have 3 classes for this data, so our confusion matrix is 3 X 3.
from sklearn.metrics import ConfusionMatrixDisplay
ConfusionMatrixDisplay.from_estimator(
pipe,
X_test,
y_test
);
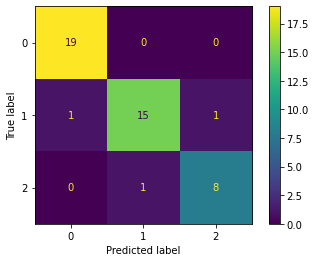
We see that we can still compute a confusion matrix, for problems with more than 2 labels in the target column.
The diagonal values are the correctly labelled wines and the rest are the errors.
Here we can see the model mistakenly predicted:
1 wine of true class 1 as class 0 and,
1 wine of true class 1 as class 2.
1 of the wines with a class of 2 as class 1.
11.4.2. Multiclass classification report#
Precision, recall, etc. don’t apply directly but like we said before, but depending on which class we specify as our “positive” label and consider the rest to be negative, then we can.
from sklearn.metrics import classification_report
print(classification_report(y_test, predictions, digits=4))
precision recall f1-score support
0 0.9500 1.0000 0.9744 19
1 0.9375 0.8824 0.9091 17
2 0.8889 0.8889 0.8889 9
accuracy 0.9333 45
macro avg 0.9255 0.9237 0.9241 45
weighted avg 0.9331 0.9333 0.9326 45
If class 0 is our positive class then our precision, recall and f1-scores are 0.95, 1.00, 0.9744 respectively.
If class 1 is our positive class then now the precision, recall and f1-scores are 0.9375, 0.8824, 0.9091.
And finally, if class 2 is our positive class then the precision, recall and f1-scores are 0.8889, 0.8889, 0.8889.
Again the support column on the right shows the number of examples of each wine class.
11.5. Multi-class coefficients#
Let’s look at the coefficients with this multi-class problem. (Ignore the max_iter for now. You can look into it here if you like)
pipe.named_steps['logisticregression'].coef_
array([[ 0.65224456, 0.15226257, 0.39849154, -0.65779374, 0.14809695,
0.40339704, 0.64196157, -0.17781306, -0.08015819, 0.14511694,
0.10843067, 0.57387779, 1.04929389],
[-0.98152274, -0.46358731, -0.72783023, 0.39233462, -0.37572384,
-0.12216444, 0.26882969, 0.28402296, 0.57145264, -0.7940844 ,
0.64433443, 0.0136787 , -1.02136195],
[ 0.32927817, 0.31132474, 0.32933868, 0.26545912, 0.22762689,
-0.2812326 , -0.91079126, -0.1062099 , -0.49129445, 0.64896746,
-0.75276509, -0.58755649, -0.02793194]])
pipe.named_steps['logisticregression'].coef_.shape
(3, 13)
What is going on here?
Well, now we have one coefficient per feature per class.
The interpretation is that these coefficients contribute to the prediction of a certain class.
The specific interpretation depends on the way the logistic regression is implementing multi-class (OVO, OVR).
11.6. Multi-class and predict_proba#
If we look at the output of predict_proba you’ll also see that there is a probability for each class and each row adds up to 1 as we would expect (total probability = 1).
pipe.predict_proba(X_test)[:5]
array([[9.93054789e-01, 6.37825640e-03, 5.66954156e-04],
[8.88767485e-04, 9.91007367e-01, 8.10386504e-03],
[9.99631873e-01, 2.80846843e-04, 8.72802725e-05],
[4.31126532e-04, 3.74655335e-04, 9.99194218e-01],
[1.36846522e-03, 9.97552882e-01, 1.07865245e-03]])
11.7. Let’s Practice#
1. Which wrapper is more computationally expensive?
2. Name a model that can handle multi-class problems without any issues or needing any additional strategies.
3. If I have 6 classes, how many models will be built if I use the One-vs-Rest strategy?
4. If I have 6 classes, how many models will be built if I use the One-vs-One strategy?
Use the diagram below to answer the next few questions:
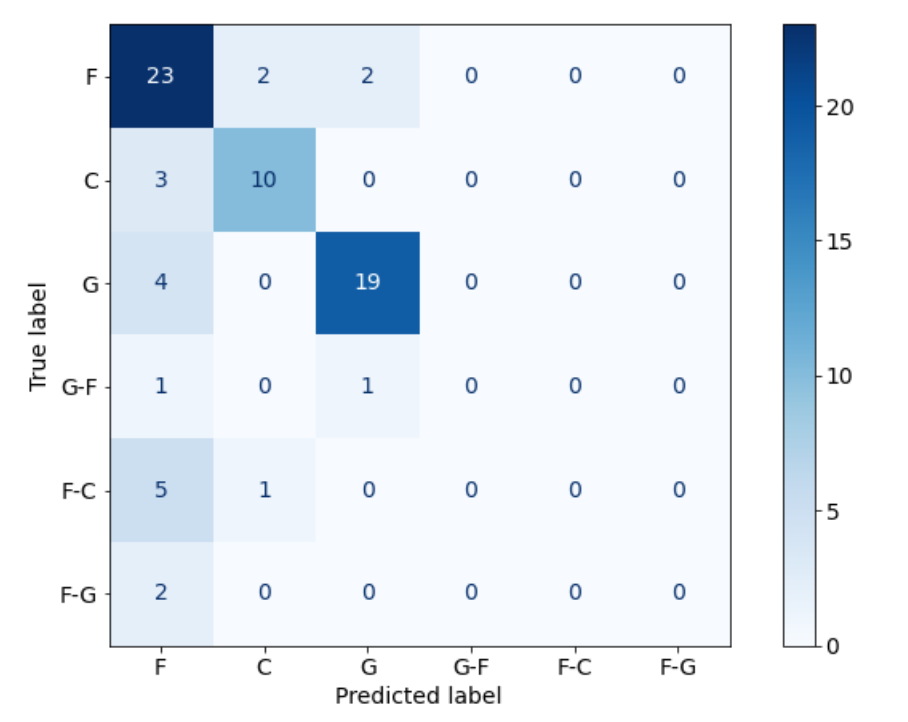
5. How many examples did the model correctly predict?
6. How many examples were incorrectly labelled as G?
7. How many F-C labels were in the data?
True or False:
8. Decision Trees use coefficients for multi-class data.
9. Using 1 target label as the positive class will make all other target labels negative.
Solutions!
One-vs-One
Decision Trees, K-nn
6
\(6(5)/2=15\)
52
3
6
False
True
11.8. Pandas Profiler#
EDA secret! (Careful to only use this on your training split though -> Golden Rule!)
quickly generate summaries of dataframes including dtypes, stats, visuals, etc.
Pandas profiling is not part of base Pandas
If using conda, install with:
pip install pandas-profiling
from pandas_profiling import ProfileReport
df = pd.read_csv('data/housing.csv')
profile = ProfileReport(df)
profile
11.9. What We’ve Learned Today#
How to carry out multi-class classification.
How to utilize pandas profiling for EDA.
